Non-CpG DNA methylation in animal genomes
Check out our new review led by Thirsa Brethouwer, on the topic of the somewhat enigmatic non-CpG methylation animal genomes, just out in Nature Genetics. Another great collaboration with the Mendoza lab.
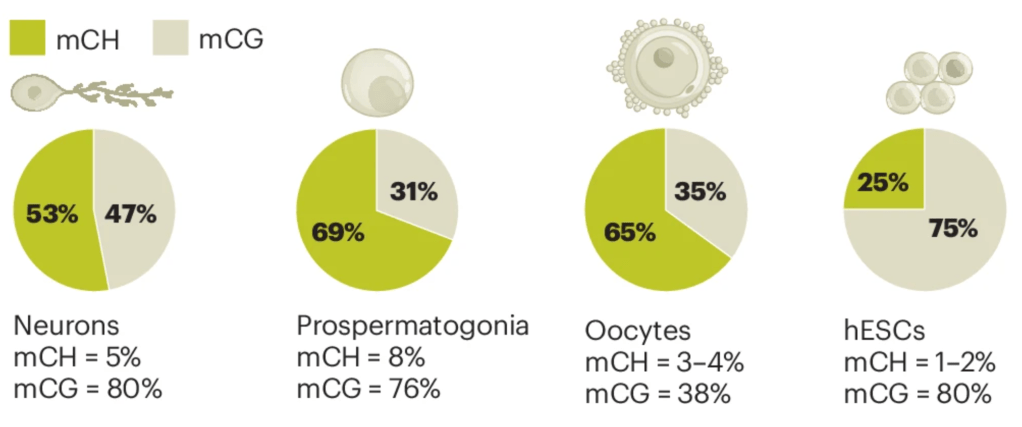
Check out our new review led by Thirsa Brethouwer, on the topic of the somewhat enigmatic non-CpG methylation animal genomes, just out in Nature Genetics. Another great collaboration with the Mendoza lab.

DANIO-ReCODE MSCA Funded
The DANIO-ReCODE Doctoral Network is funded! Ozren bogdanovic will act as the coordinator of the MSCA DN project: “DANIO-CODE – the next frontier: Decoding transcription regulation in regeneration by advanced genomics and computational tools (DANIO-ReCODE)“. The principal objective of DANIO-ReCODE is to provide world-class doctoral training to a new generation of early-career researchers interested in understanding the complex and multi-layered process of vertebrate tissue regeneration. DANIO-ReCODE will combine the multidisciplinary expertise of 15 research laboratories in EU and UK to: i) exploit the regenerative capacity of the highly tractable zebrafish (Danio rerio) model system to unravel the molecular mechanisms of heart, brain, and eye regeneration; ii) understand how regulation of regeneration in zebrafish compares to other vertebrates, including mammals; iii) identify vertebrate shared pathways for targeting regenerative therapies in humans, and iv) through the integration of state-of-the-art massively parallel sequencing technologies, genome editing, computational genomics, artificial intelligence (AI), and data visualisation, generate next generation regulatory genomics resources, which will enhance the understanding of vertebrate regenerative processes, while providing new avenues for the repair or replacement of damaged or diseased cells, organs, or tissues.

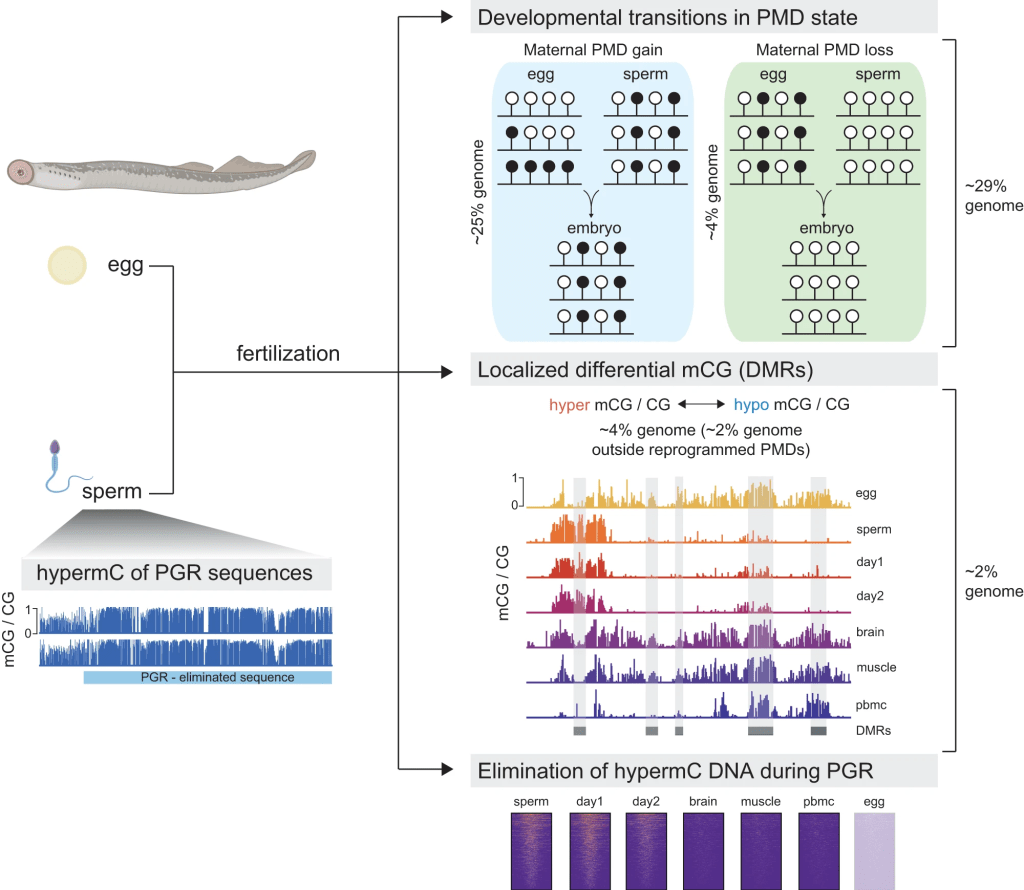
Extensive DNA methylome rearrangement during early lamprey embryogenesis
Allegra Angeloni’s first, first author paper is out in Nature Communications! Here, we studied 5mC dynamics during the embryonic development of sea lamprey, a jawless vertebrate which occupies a critical phylogenetic position as the sister group of the jawed vertebrates. We employed 5mC quantification in lamprey embryos and tissues, and discovered large-scale maternal-to-paternal epigenome remodeling that affects ~30% of the embryonic genome and is predominantly associated with partially methylated domains. We further demonstrated that sequences eliminated during programmed genome rearrangement (PGR), are hypermethylated in sperm prior to the onset of PGR. Our study thus unveils important insights into the evolutionary origins of vertebrate 5mC reprogramming, and how this process might participate in diverse developmental strategies.
Evolutionary conservation of embryonic DNA methylome remodelling in distantly related teleost species
We are very excited to see the last chapter of Sam Ross’ PhD thesis finally in press in Nucleic Acids Research. Here, we investigated the evolutionary conservation and divergence of DNA methylation remodelling in teleosts by generating base resolution DNA methylome datasets of developing medaka and medaka-zebrafish hybrid embryos. In contrast to previous reports, we show that medaka display comparable DNA methylome dynamics to zebrafish with high gametic mCG levels (sperm: ∼90%; egg: ∼75%), and adoption of a paternal-like methylome during early embryogenesis, with no signs of prior DNA methylation erasure. We also demonstrate that non-canonical DNA methylation (mCH) reprogramming at TGCT tandem repeats is a conserved feature of teleost embryogenesis. Lastly, we find remarkable evolutionary conservation of DNA methylation remodelling patterns in medaka-zebrafish hybrids, indicative of compatible DNA methylation maintenance machinery in far-related teleost species. Overall, these results suggest strong evolutionary conservation of DNA methylation remodelling pathways in teleosts, which is distinct from the global DNA methylome erasure and reestablishment observed in mammals.

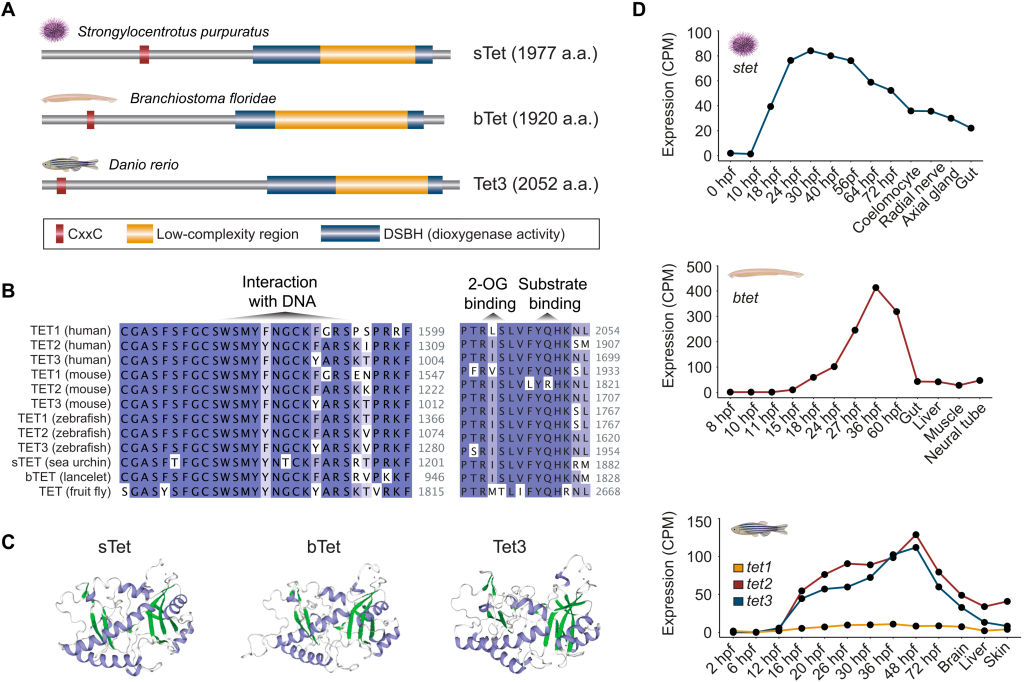
Active DNA demethylation of developmental cis-regulatory regions predates vertebrate origins
Our latest research lead by Ksenia Skvortsova is out in Science Advances. Check out this fun collaborative effort where we unravel the usage of active DNA demethylation pathways in non-vertebrate lineages. We find that invertebrate deuterostomes such as lancelet and se a urchin use TET enzymes for targeted demethylation of regulatory regions associated with developmental genes and show that the complement of identified 5hmC-regulated genes is conserved to vertebrates. This work demonstrates that active 5mC removal from regulatory regions is a common feature of deuterostome embryogenesis suggestive of an unexpected deep conservation of a major gene-regulatory module.
Zebrafish multiomics atlas
Our most recent collaborative endeavour has been published in Nature Genetics. DANIO-CODE is an international collaborative effort that aims to annotate the functional elements of the zebrafish genome. Zebrafish are increasingly used to successfully model human disease, to screen drugs and to study environmental toxicity in addition to being a model for embryonic development. Zebrafish genome is the third most complete but further annotation is essential to better realize the power of this model organism in diverse areas of biomedical research and to further strengthen its role in biomedical research. DANIO-CODE partners work together to provide a central resource of publicly available data and genome annotation of the zebrafish genome. DANIO-CODE combines a large variety of genome wide datasets including protein-coding and non-coding transcribed genome elements, non-coding functional elements such as cis-regulatory modules and associated epigenetic features. DANIO-CODE was established at a workshop held in Imperial College London in December 2014 (see meeting report published in Zebrafish) where members of the zebrafish genomics research community and previous contributors to ENCODE and FANTOM genome annotation projects identified key aims.

Lab move
The lab has recently relocated from the Garvan Institute / UNSW (Sydney, Australia) to the Andalusian Centre for Developmental Biology – CABD (Seville, Spain). We will soon be hiring at all levels. In the meantime please direct any informal inquiries to Ozren Bogdanovic (obog@upo.es).
Transient triple TET (tet1/2/3/) knockouts in zebrafish
In our latest work, Sam Ross provides a detailed protocol for the generation of triple TET zebrafish CRISPants (F0 CRISPR/Cas9 KOs) and their subsequent molecular analyses by reduced representation bisulfite sequencing (RRBS). The full protocol can be accessed here, while the entire book (TET proteins and DNA demethylation, Methods and Protocols) can be obtained through the Springer website.

TET proteins and DNA Demethylation
Check out the latest book on protocols in the field of TET proteins and DNA demethylation coedited by Michiel Vermeulen (Radboud University) and myself. The book is divided in five parts. Part One describes technologies aimed at detecting and quantifying DNA methylation turnover using massively parallel sequencing, ELISA, and mass spectrometry approaches. Part Two looks at data analyses protocols for distinguishing acting versus passive DNA demethylation and estimation of 5mC and 5hmC levels. Part Three deals with a new topic that takes advantage of modified CRISPR/Cas9 genome editing systems to target DNA demethylation activity to genomic loci of interest. Part Four discusses protocols that detail how to purify TET proteins and unravel their protein interactions, and Part Five looks at the assessment of TET protein function and activity in vivo and in vitro. Written in the highly successful Methods in Molecular Biology series format, chapters include introductions to their respective topics, lists of the necessary materials and reagents, step-by-step, readily reproducible laboratory protocols, and tips on troubleshooting and avoiding known pitfalls.
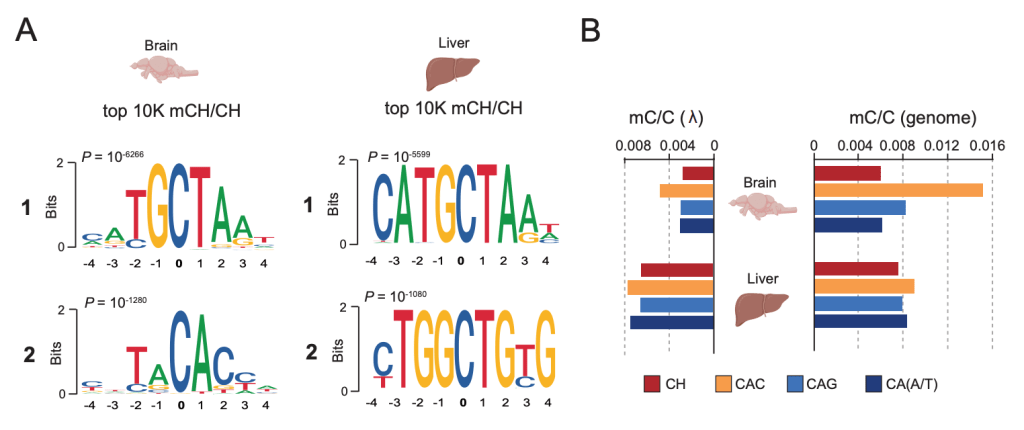
Non-CpG Methylation in the Zebrafish Brain
Our latest is out in Frontiers in Cell and Developmental Biology. Here we demonstrate that zebrafish, just like mammals, accumulate non-canonical DNA methylation (mCH) during brain development. Moreover, we identify accumulation of mCH at transposons from the Tc1-mariner superfamily. Overall, this work demonstrates the evolutionary conservation of developmental mCH dynamics and highlights the potential of zebrafish as a model to study mCH regulation and function during normal and perturbed development (i.e Rett syndrome).